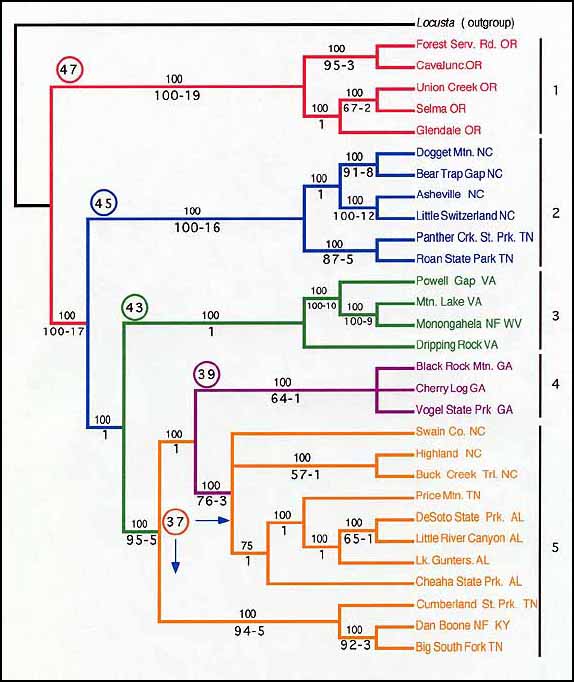
Figure 1. A consensus of 8 equally parsimonious trees
for samples of Cryptocercus from the eastern United States, based
on DNA sequence portions of the mitochondrial 12S and 16S rRNA genes and
equal weighting of characters. The numbers above branches are the percent
of times a relationship was recovered in the 8 parsimonious trees. Numbers
below branches are bootstrap values (in %) and decay indices (in steps),
respectively. Bootstrap values below 50% and decay indices of 0 are not
shown. Numbers enclosed in circles are male chromosome numbers as reported
by Kambhampati et al. (1996). Tree length: 683 steps, consistency index:
0.76, retention index: 0.84, rescaled consistency index: 0.64. (1: Cryptocercus
clevelandi, 2: Cryptocercus wrighti, 3: Cryptocercus
punctulatus, 4: Cryptocercus garciai, 5: Cryptocercus
darwini.)